Statistiek I
Multiple regression and Cronbach’s alpha
Martijn Wieling
Question 1: last lecture
Last lecture
- Simple linear regression with a nominal variable
- Multiple linear regression with multiple independent variables
- Introduction to multiple linear regression with an interaction
This lecture
- Part I: Multiple regression with an interaction (continued)
- Part II: Cronbach’s alpha to assess the reliability of questionnaires
- Part III: Recap of all lectures (time permitting)
Part I: Dataset for multiple regression
- English L2 phonetically transcribed pronunciation data from Speech Accent Archive
- Goal: identify potential determinants of L2 speakers’ English pronunciation quality
- Nativelikeness measured by comparing pronunciations to American English speakers
- Here: data from 325 L2 non-Indo-European speakers of English
- We assess the effect of age and length of residence in an English-speaking country
- Age: first as nominal variable (young vs. old), later as numerical variable
- Note that we don’t specify hypotheses here, as we conduct an exploratory analysis
- We aim to identify potentially “interesting” variables, which may serve to inform future hypotheses and data collection efforts
- For simplicity, we (wrongly) ignore variability linked to native language and country
- Mixed-effects regression is required for this: covered in Statistiek II
Investigating the influence of length of residence
Call:
lm(formula = NL ~ LR, data = saa)
Residuals:
Min 1Q Median 3Q Max
-2.9902 -0.6875 0.0454 0.6867 2.2630
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.15505 0.06455 -2.40 0.017 *
LR 0.02041 0.00466 4.38 1.6e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.973 on 323 degrees of freedom
Multiple R-squared: 0.0561, Adjusted R-squared: 0.0531
F-statistic: 19.2 on 1 and 323 DF, p-value: 1.61e-05Question 2
Adding a second variable in a multiple regression model
Call:
lm(formula = NL ~ LR + AgeGroup, data = saa)
Residuals:
Min 1Q Median 3Q Max
-3.0168 -0.6580 0.0846 0.6557 2.2357
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.12920 0.06498 -1.99 0.048 *
LR 0.02770 0.00554 5.00 9.3e-07 ***
AgeGroupOld -0.37194 0.15523 -2.40 0.017 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.966 on 322 degrees of freedom
Multiple R-squared: 0.0726, Adjusted R-squared: 0.0668
F-statistic: 12.6 on 2 and 322 DF, p-value: 5.38e-06Model comparison: additional predictor necessary?
Analysis of Variance Table
Model 1: NL ~ LR
Model 2: NL ~ LR + AgeGroup
Res.Df RSS Df Sum of Sq F Pr(>F)
1 323 306
2 322 300 1 5.36 5.74 0.017 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- The additional model complexity is supported by the improved fit to the data
Which variable is most important?
saa$LR.z <- (saa$LR - mean(saa$LR)) / sd(saa$LR)
summary(m2 <- lm(NL ~ LR.z + AgeGroup, data=saa)) # LR has larger effect (0.32 per SD; AG: 0.37 in total)
Call:
lm(formula = NL ~ LR.z + AgeGroup, data = saa)
Residuals:
Min 1Q Median 3Q Max
-3.0168 -0.6580 0.0846 0.6557 2.2357
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0813 0.0634 1.28 0.201
LR.z 0.3214 0.0642 5.00 9.3e-07 ***
AgeGroupOld -0.3719 0.1552 -2.40 0.017 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.966 on 322 degrees of freedom
Multiple R-squared: 0.0726, Adjusted R-squared: 0.0668
F-statistic: 12.6 on 2 and 322 DF, p-value: 5.38e-06Interpretation of intercept in the regression model
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.081 0.063 1.3 2.0e-01
LR.z 0.321 0.064 5.0 9.3e-07
AgeGroupOld -0.372 0.155 -2.4 1.7e-02- Young people (
AgeGroup == 'Young') with an avg. length of residence (LR.z == 0) have a predicted nativelikeness of 0.081
Interpreting the regression model logically
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.081 0.063 1.3 2.0e-01
LR.z 0.321 0.064 5.0 9.3e-07
AgeGroupOld -0.372 0.155 -2.4 1.7e-02- Summary shows \(\beta_\textrm{LR.z}\) = 0.32
- For every increase of LR of 1 SD, the nativelikeness score increases by 0.32
- Summary shows \(\beta_\textrm{AgeGroupOld}\) = -0.37
- Older speakers have a nativelikeness score that is 0.37 lower than younger speakers
- Fitted (predicted) value of the model can be determined using regression formula
- \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 \implies\)
NL = 0.08 + 0.32 * LR.z + -0.37 * AgeGroupOld AgeGroupOldequals 1 for theOldgroup and 0 for theYounggroup
- \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 \implies\)
Interpreting the regression model numerically
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.081 0.063 1.3 2.0e-01
LR.z 0.321 0.064 5.0 9.3e-07
AgeGroupOld -0.372 0.155 -2.4 1.7e-02- \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 \implies\)
NL = 0.08 + 0.32 * LR.z + -0.37 * AgeGroupOld - For
LR.zof 0 andAgeGroupYoung: 0.08 + 0.32 \(\times\) 0 + -0.37 \(\times\) 0 = 0.08 (= Intercept) - For
LR.zof 0.5 andAgeGroupOld: 0.08 + 0.32 \(\times\) 0.5 + -0.37 \(\times\) 1 = -0.13 - Note that the effects are independent and do not influence each other!
Question 3
Interpreting the regression model visually

Interaction between nominal and numerical variable
Call:
lm(formula = NL ~ LR.z * AgeGroup, data = saa)
Residuals:
Min 1Q Median 3Q Max
-2.9464 -0.6658 0.0731 0.6751 2.3044
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1386 0.0684 2.03 0.044 *
LR.z 0.5190 0.1114 4.66 4.7e-06 ***
AgeGroupOld -0.3289 0.1556 -2.11 0.035 *
LR.z:AgeGroupOld -0.2943 0.1360 -2.16 0.031 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.961 on 321 degrees of freedom
Multiple R-squared: 0.0859, Adjusted R-squared: 0.0774
F-statistic: 10.1 on 3 and 321 DF, p-value: 2.37e-06Interaction necessary to include?
Recall regression formula for interaction from last lecture
- Regression formula \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 + \beta_{1,2} x_1 x_2 + \epsilon\)
- \(y\): dependent variable (= predicted or fitted values), \(x_i\): independent variables, \(\epsilon\): resid.
- \(\beta_0\): intercept (value of \(y\) when all \(x_i's\) are \(0\))
- \(\beta_1\): influence (slope) of \(x_1\) on \(y\) when \(x_2\) equals 0
- \(\beta_2\): influence (slope) of \(x_2\) on \(y\) when \(x_1\) equals 0
- \(\beta_{1,2}\): interaction effect (slope) of \(x_1\) and \(x_2\)
- \(y\): dependent variable (= predicted or fitted values), \(x_i\): independent variables, \(\epsilon\): resid.
Interpreting the interaction numerically
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.14 0.068 2.0 4.4e-02
LR.z 0.52 0.111 4.7 4.7e-06
AgeGroupOld -0.33 0.156 -2.1 3.5e-02
LR.z:AgeGroupOld -0.29 0.136 -2.2 3.1e-02- Regression formula \(y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 + \beta_{1,2} x_1 x_2
\implies\)
NL = 0.14 + 0.52 * LR.z + -0.33 * AGOld + -0.29 * LR.z * AGOld- For
LR.zof 0 andAgeGroupYoung: 0.14 + 0.52\(\times\)0 + -0.33\(\times\)0 + -0.29\(\times\)0\(\times\)0 = 0.14 - For
LR.zof 0 andAgeGroupOld: 0.14 + 0.52\(\times\)0 + -0.33\(\times\)1 + -0.29\(\times\)0\(\times\)1 = -0.19 - For
LR.zof 0.5 andAgeGroupOld: 0.14 + 0.52\(\times\)0.5 + -0.33\(\times\)1 +-0.29\(\times\)0.5\(\times\)1 = -0.075
- For
Question 4
Interpreting the interaction logically
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.14 0.068 2.0 4.4e-02
LR.z 0.52 0.111 4.7 4.7e-06
AgeGroupOld -0.33 0.156 -2.1 3.5e-02
LR.z:AgeGroupOld -0.29 0.136 -2.2 3.1e-02- For the
YoungAgeGroup, each unit increase ofLRincreasesNLby 0.52 - For the
OldAgeGroup, each unit increase ofLRincreasesNLby 0.52 + -0.29 = 0.23 - The
LRslope is shifted downwards by 0.23 for theOldAgegroup - Summary: the effect of
LRis less beneficial for older people- Learning a language through immersion is more effective when young than old
Interpreting the interaction visually
Interaction between two nominal variables
m4 <- lm(NL ~ AgeGroup * Sex, data=saa) # We drop LR for simplicity (normally you would include it)
summary(m4) # no significant predictors
Call:
lm(formula = NL ~ AgeGroup * Sex, data = saa)
Residuals:
Min 1Q Median 3Q Max
-3.147 -0.648 0.042 0.696 2.142
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0303 0.0930 -0.33 0.74
AgeGroupOld 0.2448 0.2052 1.19 0.23
SexMale 0.0337 0.1262 0.27 0.79
AgeGroupOld:SexMale -0.3308 0.2718 -1.22 0.22
Residual standard error: 1 on 321 degrees of freedom
Multiple R-squared: 0.00546, Adjusted R-squared: -0.00384
F-statistic: 0.587 on 3 and 321 DF, p-value: 0.624- Is this model an improvement over the simpler model?
Question 5
Which model for comparison?
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.00844 0.0875 0.0965 0.923
AgeGroupOld 0.05616 0.1347 0.4171 0.677
SexMale -0.03760 0.1119 -0.3362 0.737 Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0120 0.0628 -0.191 0.849
AgeGroupOld 0.0549 0.1344 0.408 0.683 Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0200 0.0829 0.241 0.810
SexMale -0.0363 0.1117 -0.325 0.745- None of the simpler models have a significant predictor, so which one to use?
Note about model comparison for an interaction
- Note that a model with an interaction term (
A:B) should be compared to the best model without that term- This is the model including
A+B, if both terms are significant - This is the model only including
A, ifAis significant andBis not - This is the model only including
B, ifBis significant andAis not - This is the model without
AandB, if neither are significant
- This is the model including
- It is important to get this comparison right, as an interaction might provide an improvement over including
A+B(if both are not significant), but may not be an improvement over the model withoutAandB.- In that case one should stick with the model without
A,B, or their interaction
- In that case one should stick with the model without
Model comparison
m0a <- lm(NL ~ 1, data=saa) # model without AgeGroup and Sex for comparison
anova(m0a, m4) # interaction is not supportedAnalysis of Variance Table
Model 1: NL ~ 1
Model 2: NL ~ AgeGroup * Sex
Res.Df RSS Df Sum of Sq F Pr(>F)
1 324 324
2 321 322 3 1.77 0.59 0.62- In line with our guess, the interaction between
AgeGroupandSexis not supported AgeGroupis only significant if we also take into account the effect ofLR- As mentioned: in a multiple regression model the interpretation of the effect of a variable always entails that one controls for all other variables in the model
Visualization (of non-significant interaction)
Interaction between two numerical variables
saa$Age.z <- (saa$Age - mean(saa$Age)) / sd(saa$Age)
summary(m5 <- lm(NL ~ LR.z * Age.z, data=saa)) # Instead of AgeGroup we use numerical Age
Call:
lm(formula = NL ~ LR.z * Age.z, data = saa)
Residuals:
Min 1Q Median 3Q Max
-3.004 -0.670 0.116 0.670 2.237
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0714 0.0623 1.15 0.2520
LR.z 0.4763 0.0923 5.16 4.3e-07 ***
Age.z -0.1752 0.0654 -2.68 0.0078 **
LR.z:Age.z -0.1242 0.0561 -2.21 0.0277 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.959 on 321 degrees of freedom
Multiple R-squared: 0.0882, Adjusted R-squared: 0.0796
F-statistic: 10.3 on 3 and 321 DF, p-value: 1.62e-06Model comparison
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.63e-16 0.0535 -3.05e-15 1.00e+00
LR.z 3.32e-01 0.0657 5.06e+00 7.11e-07
Age.z -1.65e-01 0.0657 -2.52e+00 1.23e-02Analysis of Variance Table
Model 1: NL ~ LR.z + Age.z
Model 2: NL ~ LR.z * Age.z
Res.Df RSS Df Sum of Sq F Pr(>F)
1 322 300
2 321 295 1 4.5 4.89 0.028 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Interaction is significant
Question 6
Numerical variable better than nominal variable?
Analysis of Variance Table
Model 1: NL ~ LR.z * AgeGroup
Model 2: NL ~ LR.z * Age.z
Res.Df RSS Df Sum of Sq F Pr(>F)
1 321 296
2 321 295 0 0.724 - Both models are equally complex (same nr. of parameters), but
m5provides a better fit (RSS lower)- Take note: it’s not a good idea to “simplify” numerical variables by converting to nominal variables!
Visual interpretation of the interaction (1)

Visual interpretation of the interaction (2)
For comparison: the model without interaction
Assumptions satisfied? (1/2)
Assumptions satisfied? (2/2)
Potential solutions of not satisfying the assumptions
- Use an appropriate model
- Here we did not take into account variability associated with native language
- Mixed-effects regression is necessary for that
- Make sure all important variables are included in the model
- Transform the dependent variable (e.g., log-transformation)
- Use a generalized linear regression model (which has fewer assumptions)
- These potential solutions are not covered during this course (but in Statistiek II)
Part II: Questionnaires
- Questionnaires are an easy way to obtain much data
- But: how to ask the right question?
- For example: How do students feel about statistics?
- What type of statistics?
- What kind of feelings?
- For this reason, researchers ask several questions all aimed at acquiring similar information (i.e. questions are combined in a so-called scale)
Reliability and validity
- Validity: are you measuring what you intend to measure?
- This can be checked by consulting experts, comparing to similar measures, etc.
- Reliability: is your measure consistent?
- I.e. are results repeatable when obtained given the same conditions?
- This can be statistically assessed using Cronbach’s \(\alpha\)
- Reliability \(\neq\) validity!
Question 7
Reliability and validity
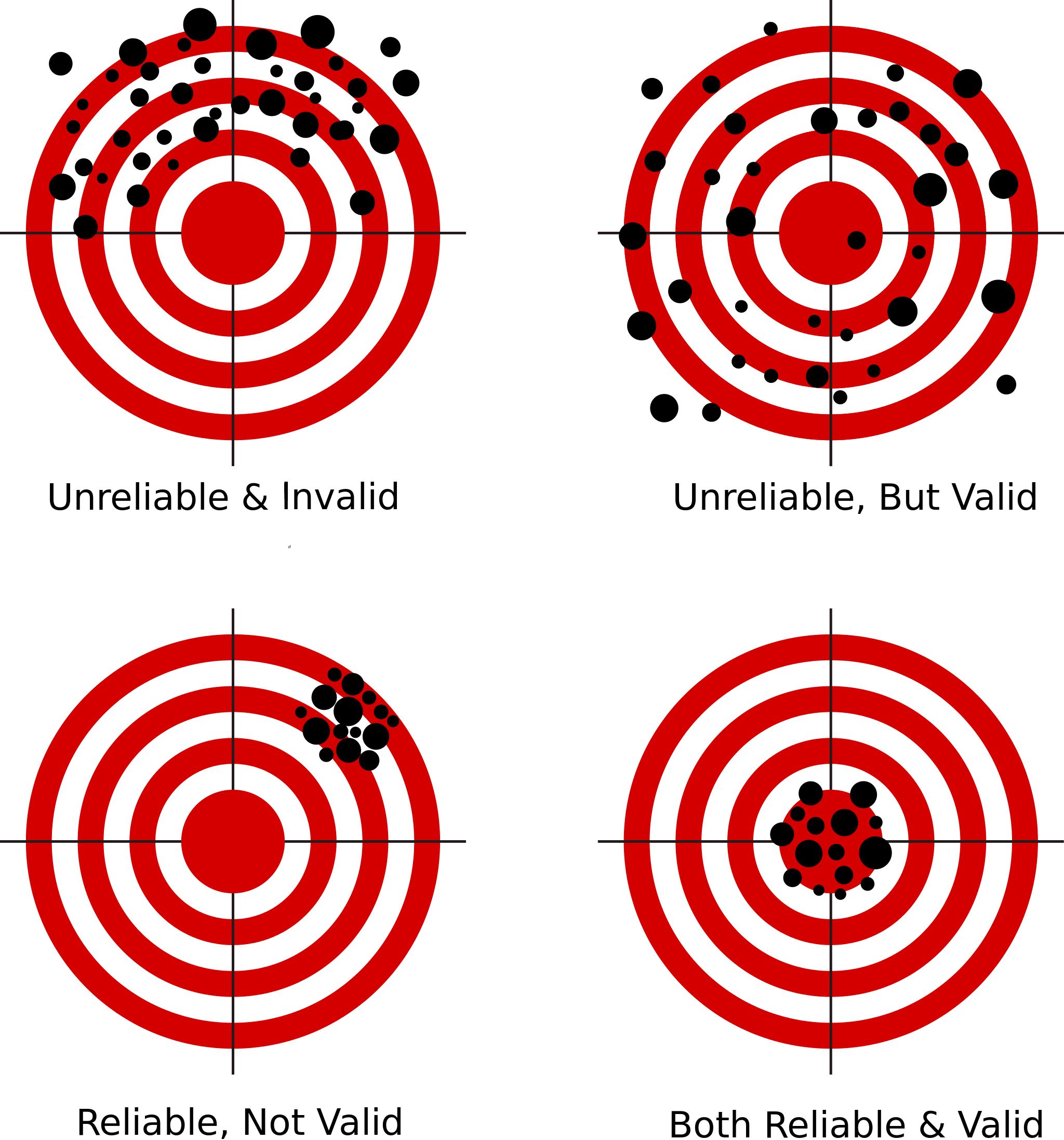
Reliability: Cronbach’s \(\alpha\) (1)
- Underlying idea:
- If we split the questions, how well do two halves of the questionnaire agree?
- And if there are many questions, how well would they agree if we looked at all possible ways of splitting?
- More information: https://www.ijme.net/archive/2/cronbachs-alpha.pdf
Reliability: Cronbach’s \(\alpha\) (2)
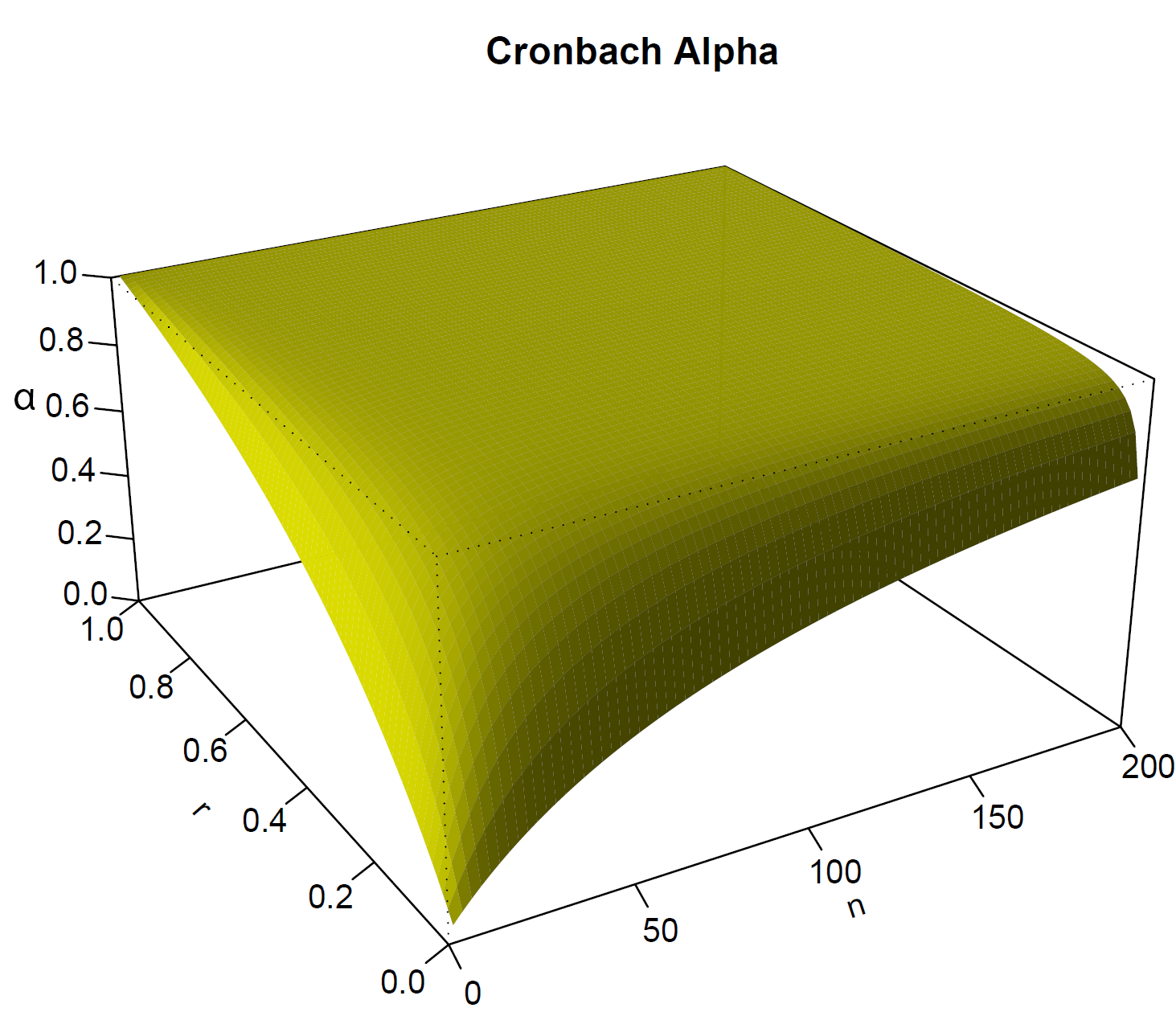
- Cronbach’s \(\alpha\) depends on:
- The average correlation \(r\) of all variables (i.e. questions) involved
- The number of questions
- Removing a problematic question may increase Cronbach’s \(\alpha\)
- Cronbach’s \(\alpha\): > 0.7 is acceptable, 0.8 is good, 0.9 is very good
- With Cronbach’s \(\alpha\) > 0.7: mean of items can be used as summary of the scale
Cronbach’s \(\alpha\) depends on \(n\) and \(r\)
Questionnaire: your feelings toward statistics
- Questions (in English): https://www.let.rug.nl/wieling/Statistiek-I/Final36scoring.pdf
Q3 Q4 Q14 Q15 Q18 Q19 Q28
1 7 2 6 2 2 6 1
2 4 4 3 4 4 4 4
3 3 3 6 6 5 2 6
4 5 4 6 4 3 6 2
5 4 5 6 4 4 2 5
6 4 2 6 2 3 5 2- Note that I have included one question which is part of a different scale
- We will try to identify this question later
Cronbach’s \(\alpha\) in R (1)
Some items ( Q3 Q19 ) were negatively correlated with the first principal component and
probably should be reversed.
To do this, run the function again with the 'check.keys=TRUE' option
Reliability analysis
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.41 0.37 0.63 0.078 0.59 0.039 4.2 0.69 0.042- Reliability is very low, but we ignored some questions having an inverted scale
Taking into account inverted questions
- The instructions of the questionnaire show that questions 4, 15, 18 and 28 should be inverted (1 \(\rightarrow\) 7, 2 \(\rightarrow\) 6, 3 \(\rightarrow\) 5, 4 \(\rightarrow\) 4, etc.)
Can we improve reliability by dropping an item?
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q3 0.74 0.69 0.73 0.27 2.18 0.02 0.10 0.36
Q4- 0.74 0.70 0.76 0.28 2.29 0.02 0.09 0.37
Q14 0.84 0.84 0.85 0.46 5.10 0.01 0.02 0.38
Q15- 0.77 0.73 0.80 0.31 2.67 0.01 0.12 0.37
Q18- 0.73 0.69 0.75 0.27 2.18 0.02 0.09 0.35
Q19 0.75 0.69 0.73 0.27 2.21 0.02 0.10 0.35
Q28- 0.71 0.67 0.73 0.26 2.06 0.02 0.08 0.35- Dropping Q14 would yield a higher Cronbach’s \(\alpha\)
Reliability without Q14
Question 8
Part III: Recap
- In the following, an overview of the contents of the previous lectures is provided
What have we covered in the past lectures? (1)
- Lecture 1: statistics and
R- Why use statistics?
- How to use R:
- Variables, functions, importing data, viewing data, modifying data, visualization, and statistics
- Lecture 2: descriptive statistics
- Four variable types
- Measures of central tendency and spread
- Standardized scores (\(z\)-scores)
- Distribution of a variable: normal distribution
What have we covered in the past lectures? (2)
- Lecture 3: sampling
- Sample vs. population
- Standard deviation for comparing individual to population
- Standard error for comparing sample to population
- Definition of \(p\)-value (probability of data given hypothesis / probability of type-I error)
- Statistical significance (\(p\)-value vs. \(\alpha\)-value)
- Reasoning about population: confidence interval
- Reasoning about population: hypothesis tests (\(H_0\) vs. \(H_a\))
- One-sided vs. two-sided hypothesis
- Comparing sample to population using standardized test: \(z\)-test
- Error types
- Sample vs. population
What have we covered in the past lectures? (3)
- Lecture 4: introduction to linear regression
- Correlation as descriptive statistic
- Simple linear regression with a single numerical predictor
- Dependent (DV) vs. independent variable (IV)
- Fitted values vs. residuals
- Assumptions: residuals normally distributed and homoscedastic, linear relationship between IV and DV
- Interpreting and visualizing output
- Effect size
- Reporting results
What have we covered in the past lectures? (4)
- Lecture 5: (multiple) linear regression
- Simple linear regression with a single nominal predictor
- Multiple linear regression
- Adding multiple independent variables
- Additional assumption: no collinearity between IVs
- Model comparison
- Determining importance of independent variables
- Interactions between two variables (introduction)
Recap
- In this lecture, we’ve covered
- Interactions in multiple regression:
- Interaction between a nominal and a numerical independent variable
- Interaction between two nominal independent variables
- Interaction between two numerical independent variables
- Cronbach’s alpha
- Interactions in multiple regression:
- Next lecture: Practice exam!
Please evaluate this lecture!
Exam question
Questions?
Thank you for your attention!
https://www.martijnwieling.nl
m.b.wieling@rug.nl





